Engineering uses science and math to solve real world problems. Cellular and Molecular Engineering uses the principles of engineering and life sciences to manipulate and understand biologic processes at cellular or molecular levels. Cells were discovered by Robert Hooke in 1665. Proteins were discovered by Antoine Fourcroy in 1789. Nucleic acids were discovered by Friedrich Miescher in 1868. In the 21st century, it is time to use principles we have learned from them to create something new, useful, and fun.
About us
Our lab has mainly focused on studying the molecular mechanisms controlling the self-renewal and cell fate determination of embryonic stem cells, especially those related to noncoding RNAs including microRNAs and long noncoding RNAs, the so called dark matter in cells. However, our current research mainly focuses on using interdisciplinary engineering approaches to develop new technologies addressing problems in RNA biology and stem cell biology. We are also interested in developing new genome editing tools. Devoted students and researchers with background in cell biology, molecular biology, biochemistry, chemistry, and bioinformatics with an ambition in creating new things to understand biology or benefit human health are always welcome to visit or join us.
Major Achievements in past 10 years
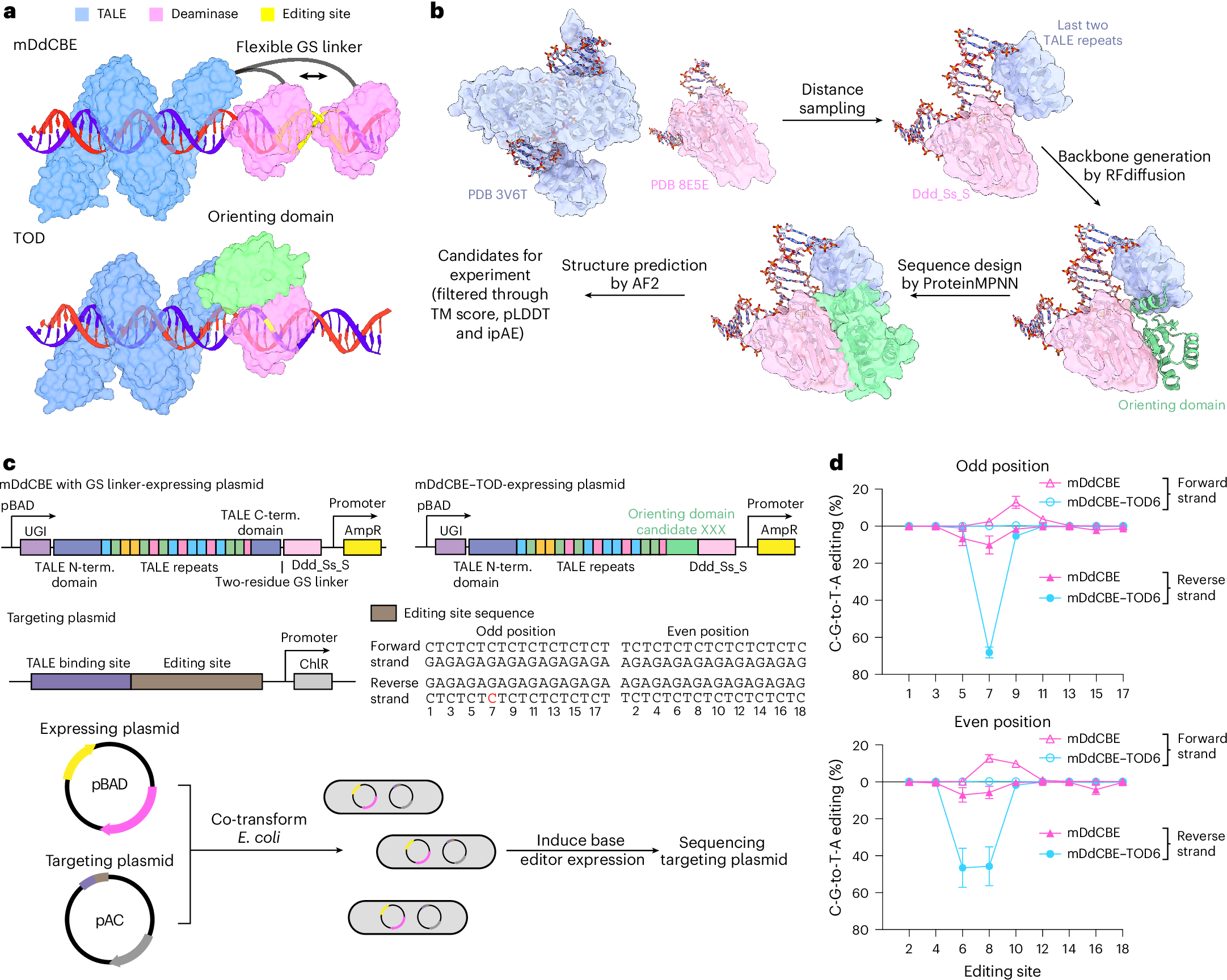
Developed a high-precision mitochondrial DNA cytosine base editor (DdCBE-TOD) based on AI-assisted protein design strategy, achieving single base precision fixed-point editing.
Mi, Li & Lv, et al. Nature Structural & Molecular Biology 2025
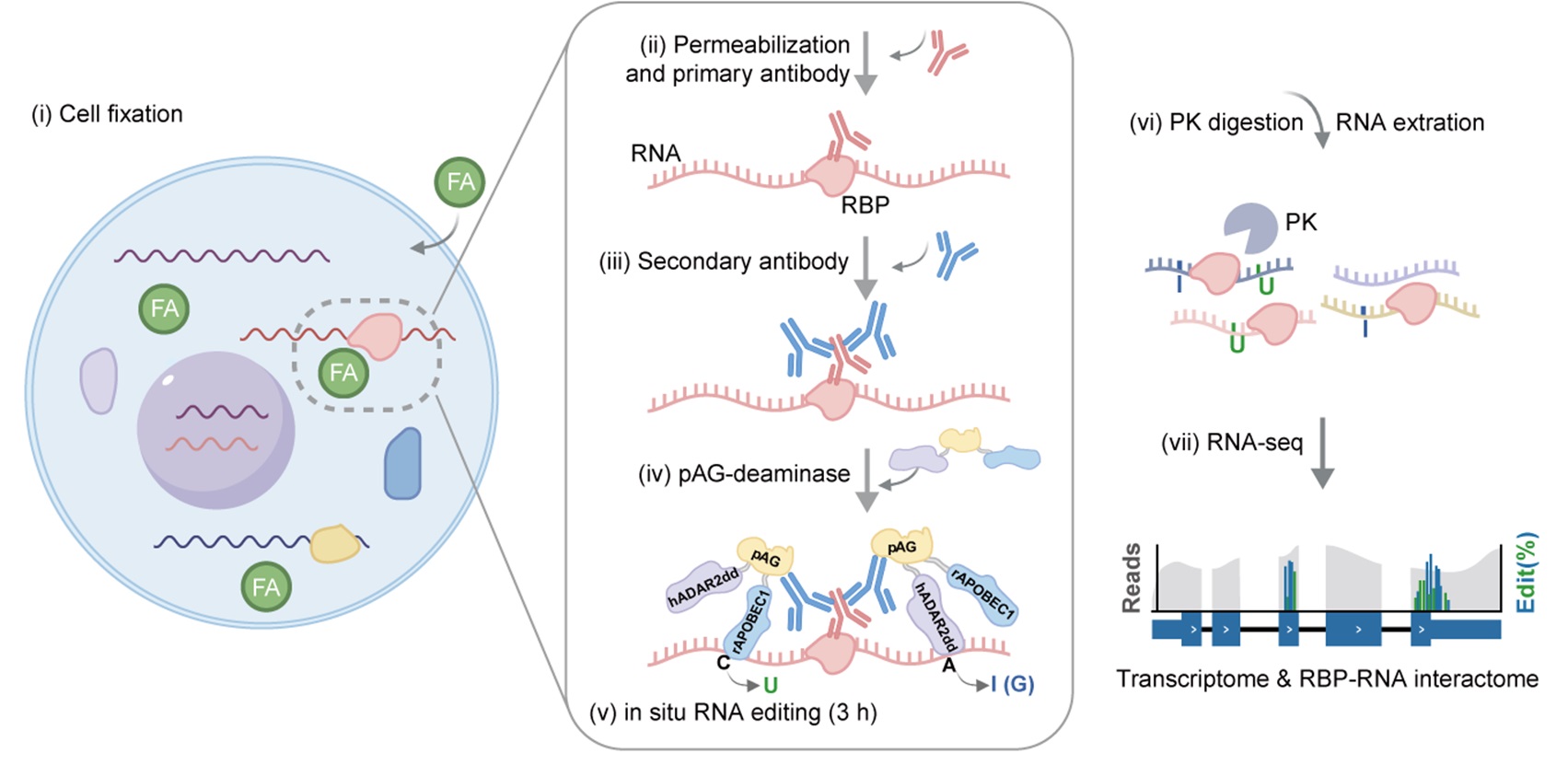
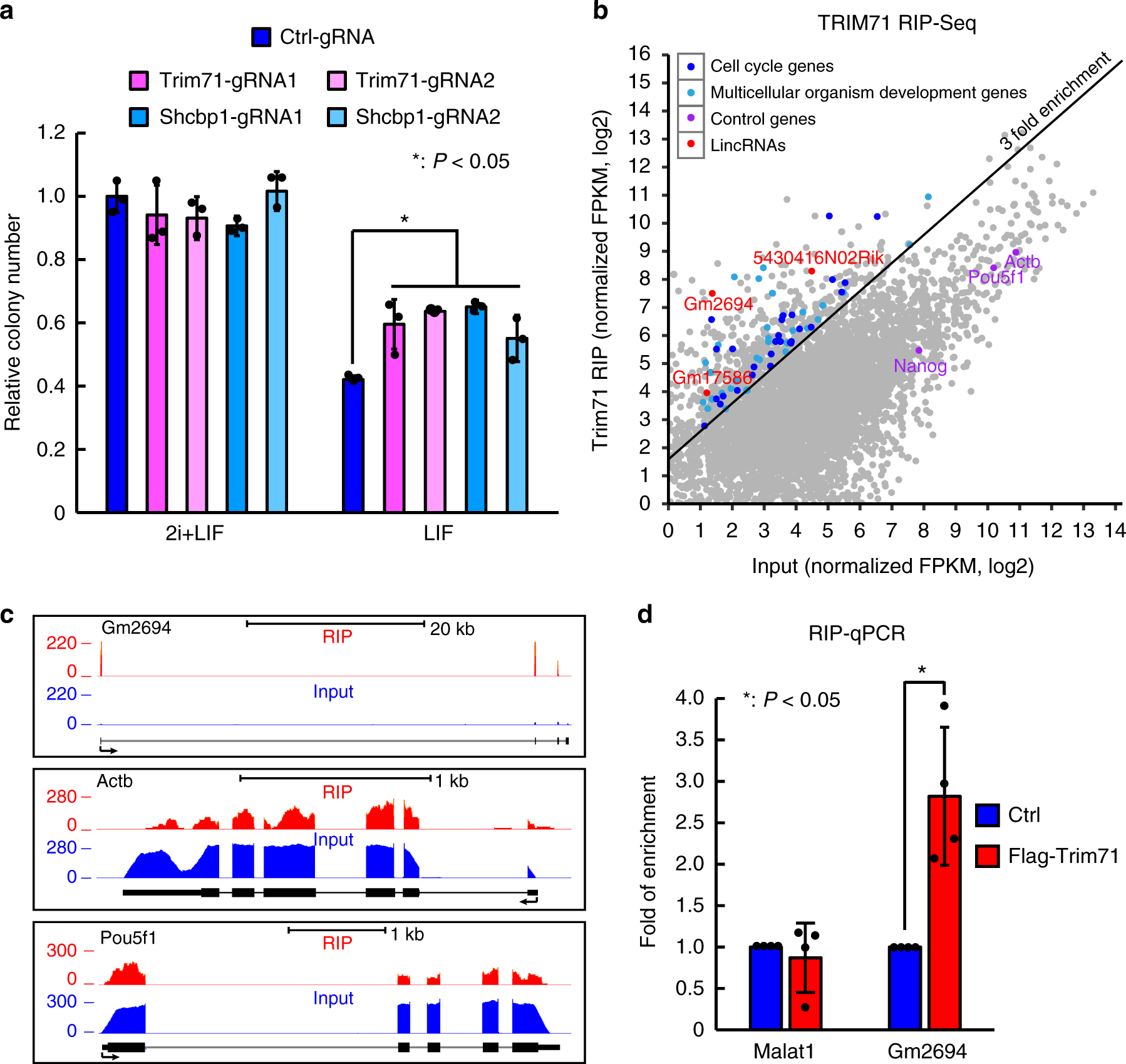
MAPIT-seq (modification added to RBP-interacting transcript sequencing): Co-profiling of in situ RNA-protein interactions and transcriptome in single cells and tissues.
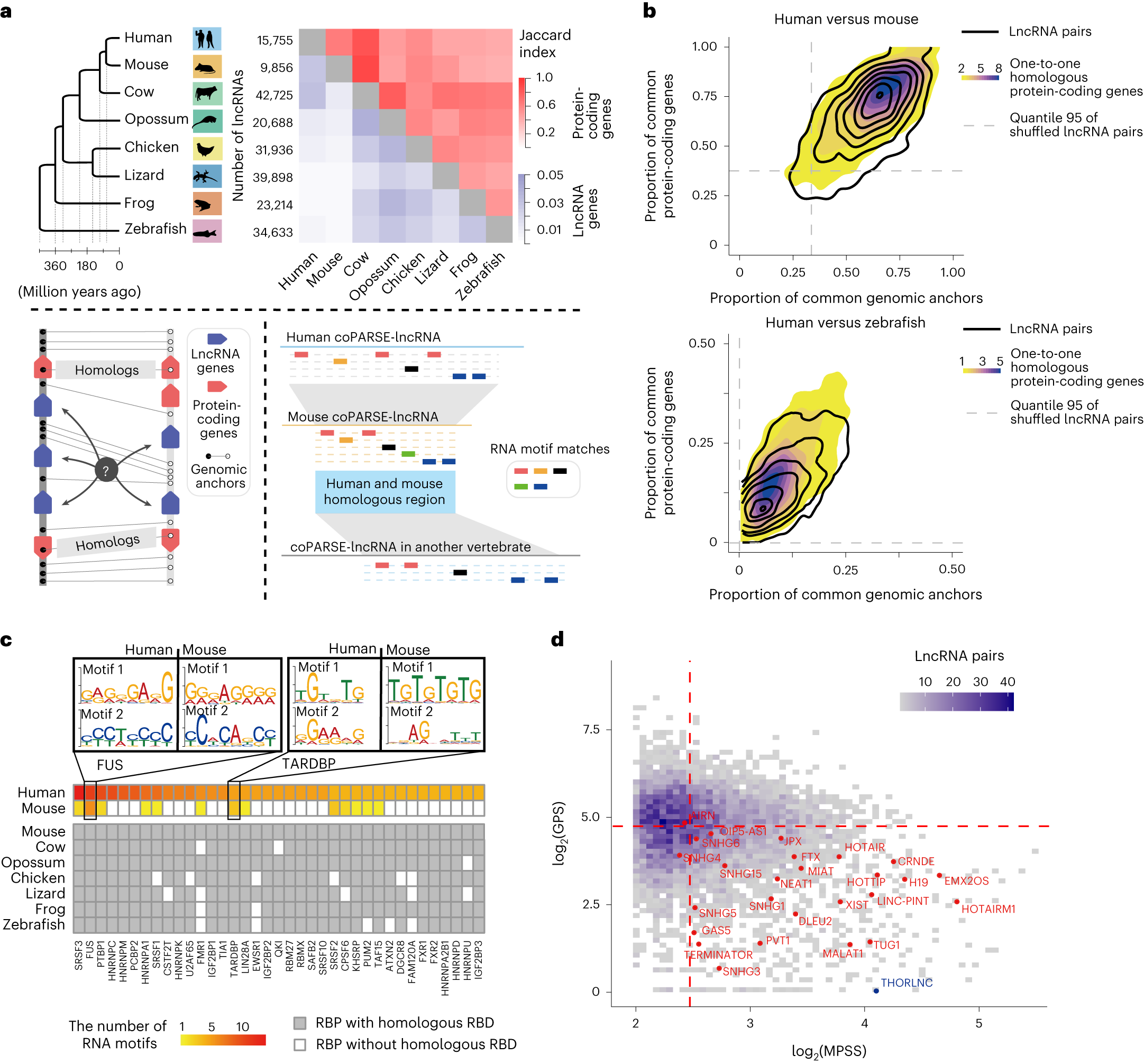
Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human.
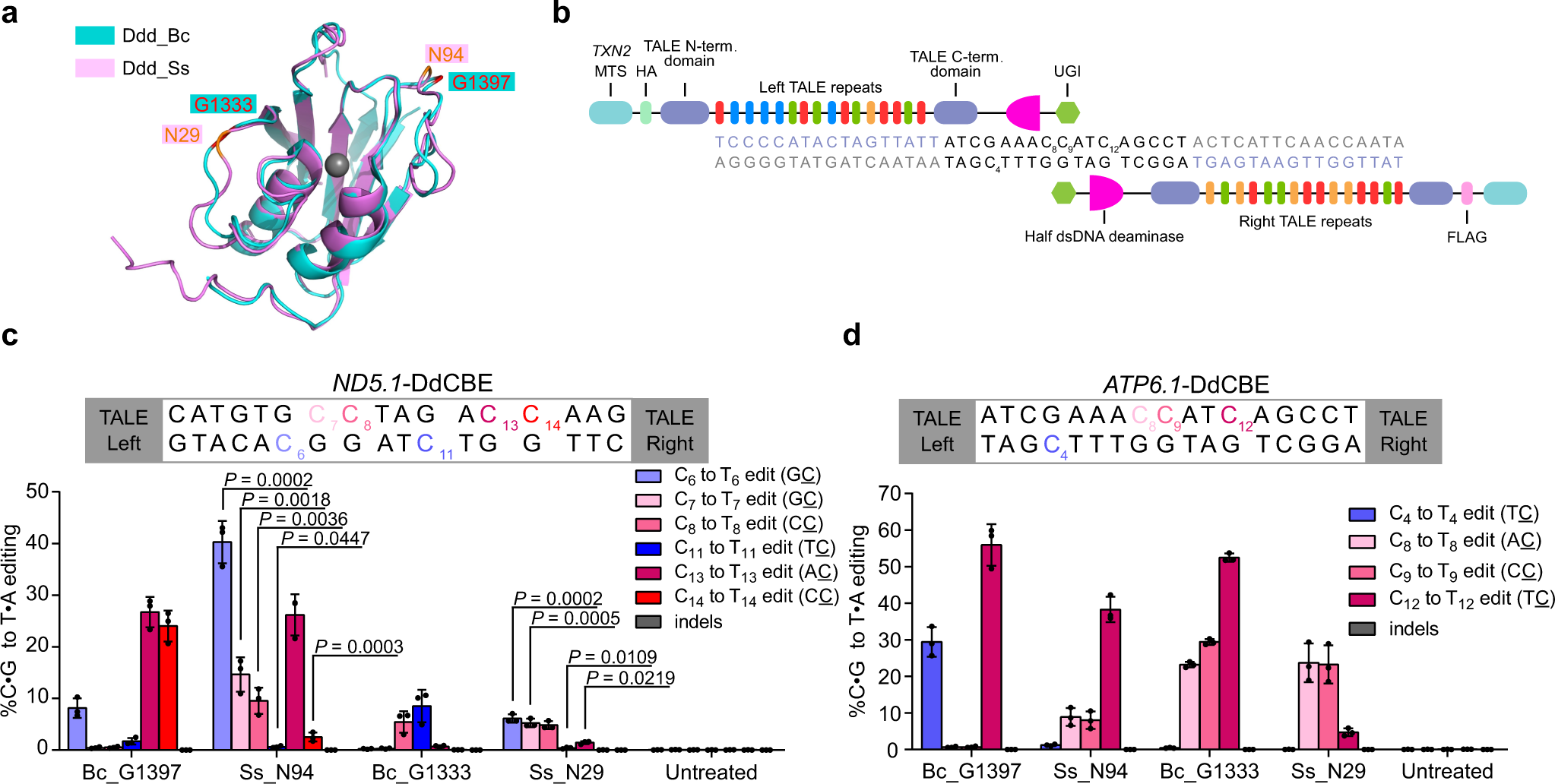
Developed Ddd_Ss-derived cytosine base editors (DdCBE_Ss) and introduced mutations at multiple mitochondrial DNA loci including previously inaccessible GC context.
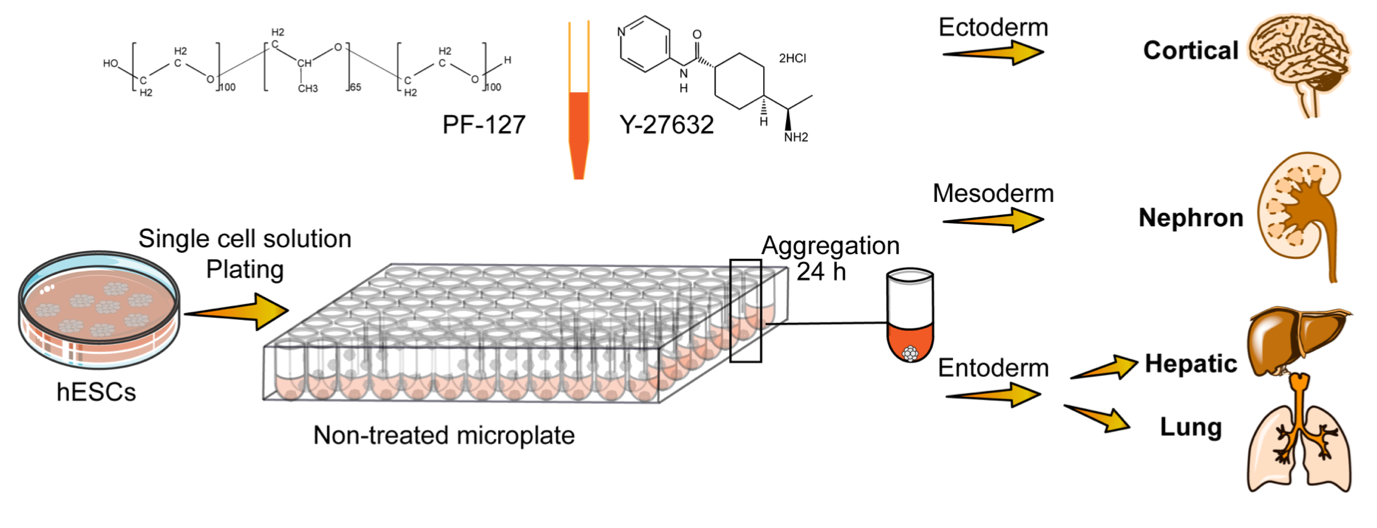
Highly reproducible and cost-effective one-pot organoid differentiation using a novel platform based on PF-127 triggered spheroid assembly.
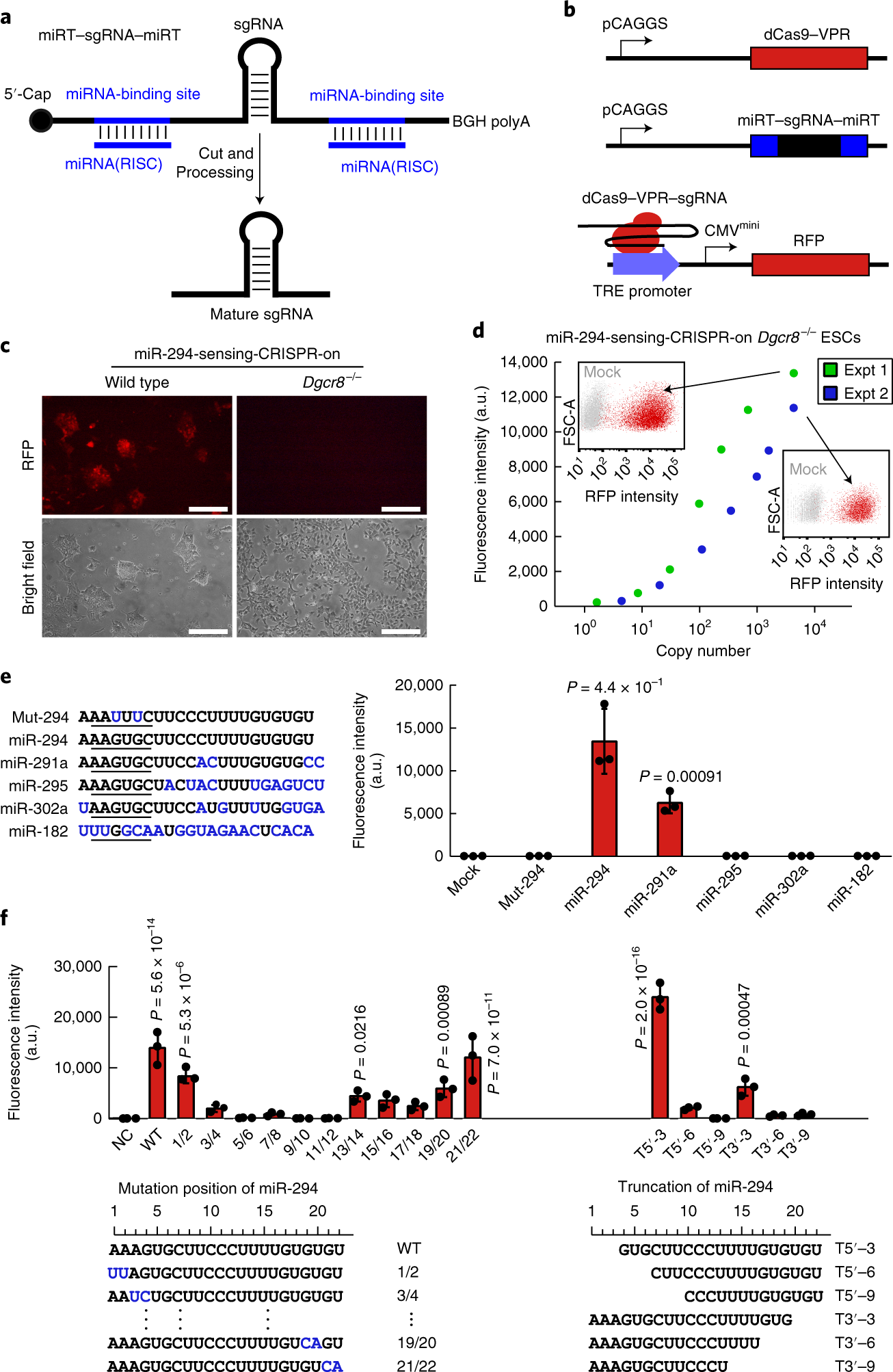
Invented miRNA inducible CRISPR-Cas9 platform (MICR) that can be used as a sensor for miRNA expression and cell-specific genome editing tool.
Discovered the novel molecular pathway, PIAS4-SUMO2-DPPA2/4, regulating zygotic genome activation (ZGA) in early embryonic development and transformation of totipotent stem cells in vitro.
Identified a lncRNA regulating FGF/ERK signaling and self-renewal of ESCs.